Selected Publications
Code, data, and PDF are provided. Ask me for questions.
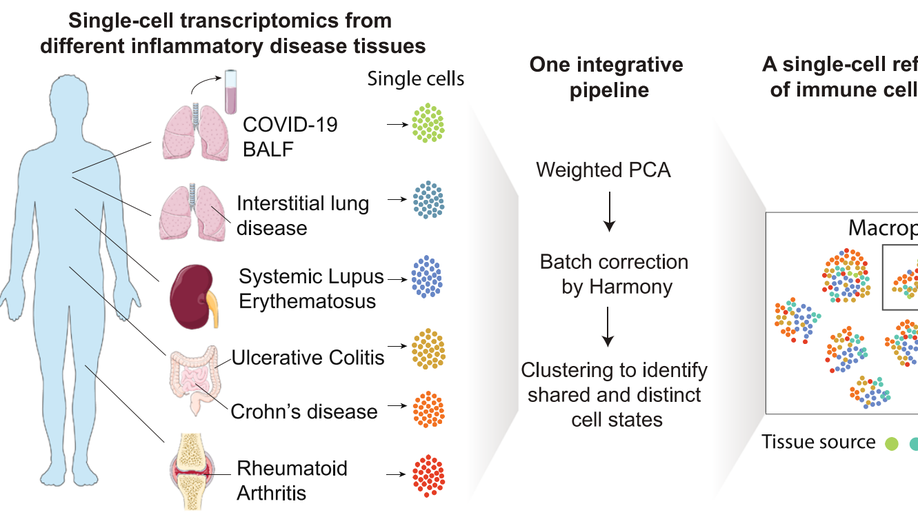
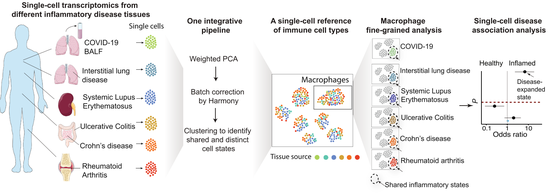
IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Medicine, 2021
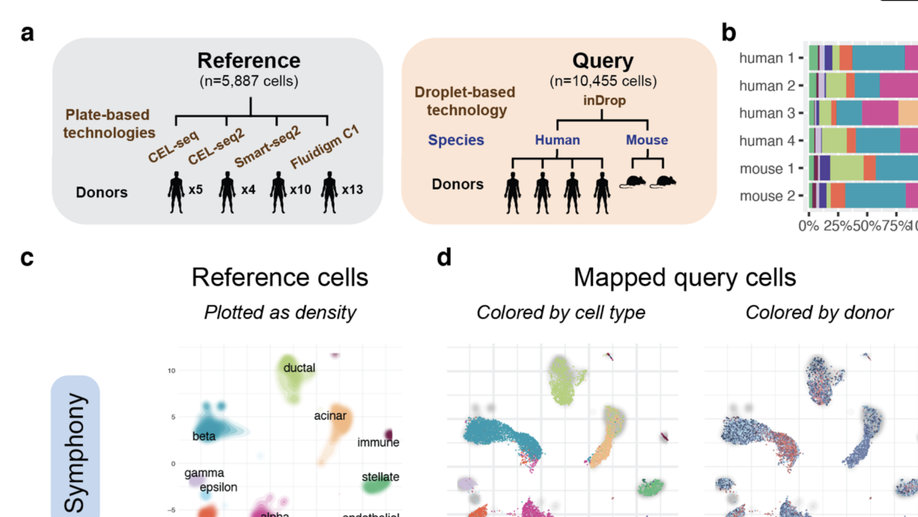
Efficient and precise single-cell reference atlas mapping with Symphony. In review, 2021
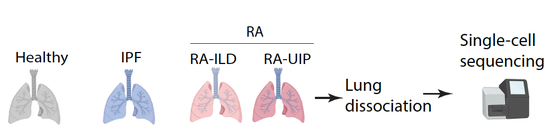
Granzyme K+ CD8 T cells (TteK) form the core population of inflamed human tissue-associated CD8 T cells. In review, 2021
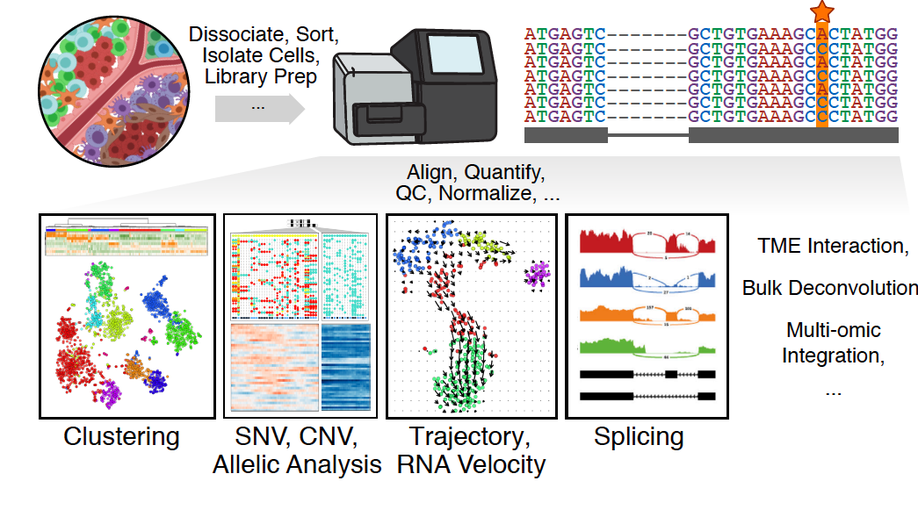
Single-cell transcriptomics in cancer: computational challenges and opportunities. Nature Experimental & Molecular Medicine, 2020
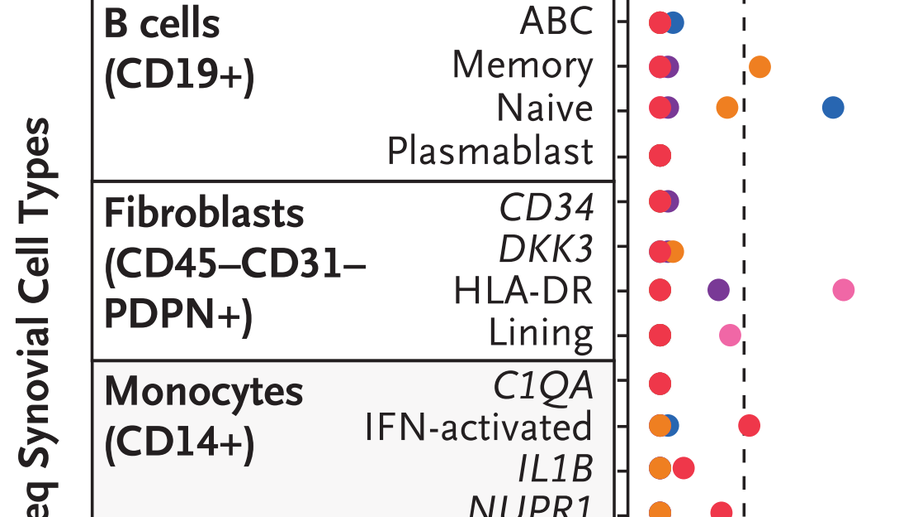
RNA Identification of PRIME Cells Predicting Rheumatoid Arthritis Flares. New England Journal of Medicine, 2020
Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Method, 2019.
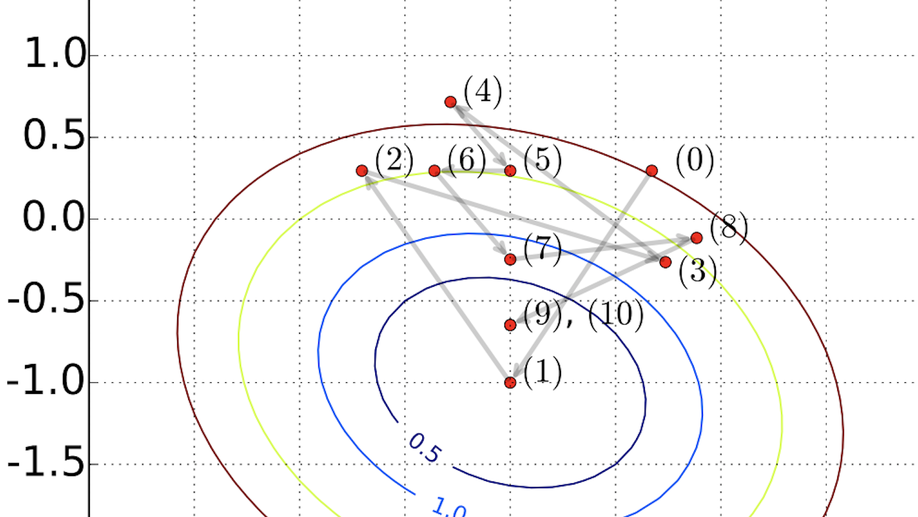
A global optimization algorithm for sparse mixed-membership matrix factorization. ICSA Book Series in Statistics. Springer, Cham, 2019
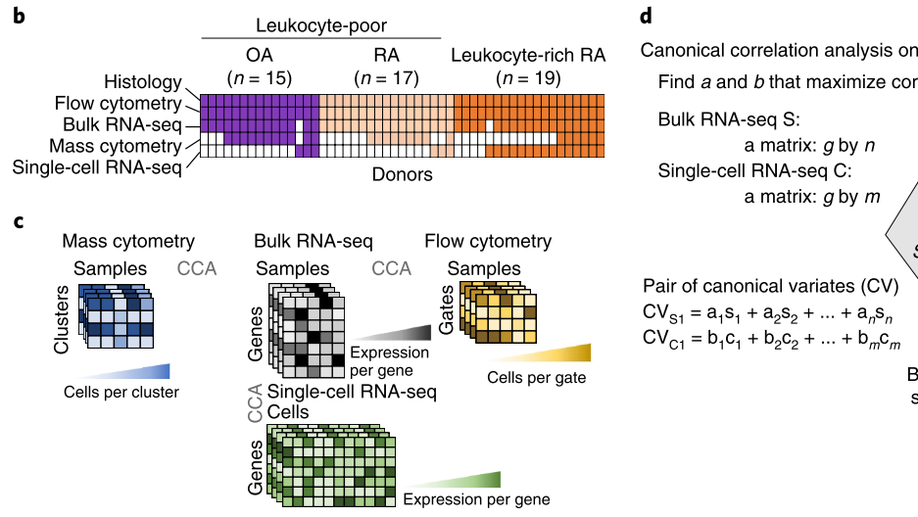
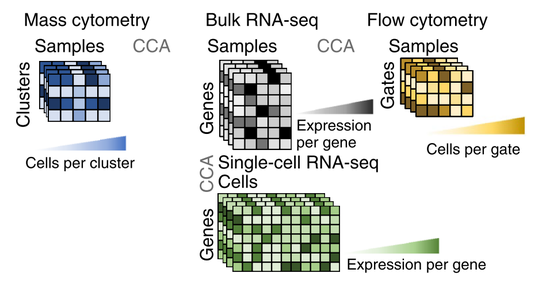
Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass sytometry. Nature Immunology, 2019.
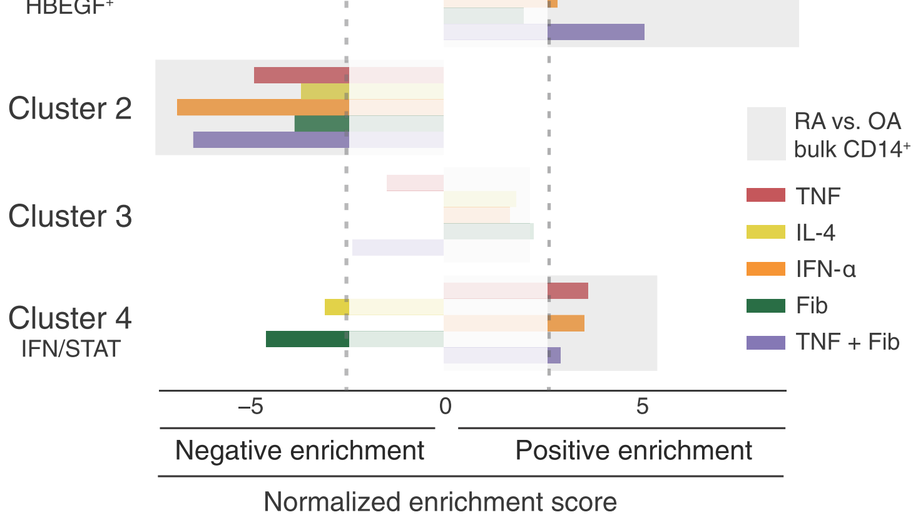
HBEGF+ macrophages identified in rheumatoid arthritis promote joint tissue invasiveness and are reshaped differentially by medications. Science Translational Medicine, 2019
The immune cell landscape in kidneys of patients with lupus nephritis. Nature Immunology, 2019
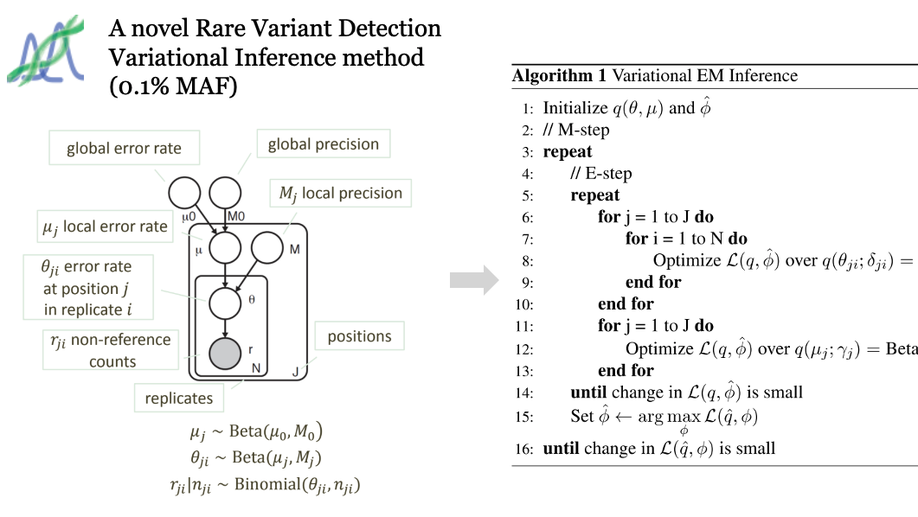
Variational inference for rare variant detection in deep, heterogeneous next generation sequencing data. BMC Bioinformatics, 2017