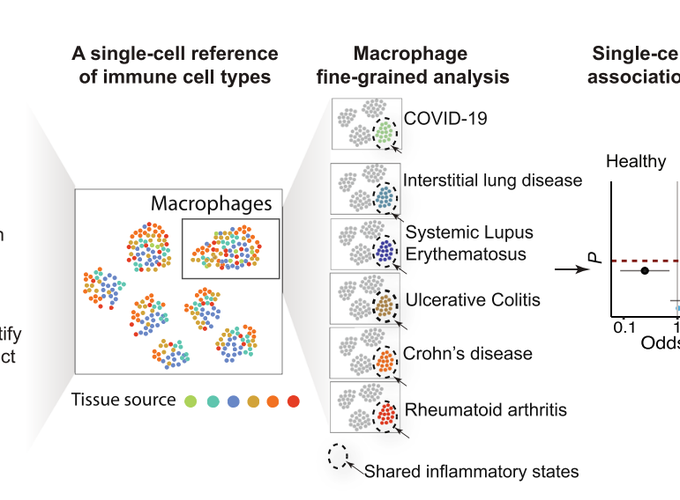
Motivation: Understanding the immune cell states shared between COVID-19 and other inflammatory diseases with established therapies may help nominate immunomodulatory therapies.
Build an immune cell reference consisting of >300,000 single-cell transcriptomic profiles from COVID-19 affected lungs and tissues from healthy subjects and patients with 5 inflammatory diseases: rheumatoid arthritis (RA), Crohn’s disease (CD), ulcerative colitis (UC), lupus, and interstitial lung disease. We tested the association of shared immune states with severe/inflamed status compared to healthy using mixed-effects modeling.
Identify a CXCL10+ CCL2+ inflammatory macrophage phenotype that is shared and strikingly abundant in severe COVID-19 bronchoalveolar lavage samples, inflamed RA synovium, inflamed CD ileum and UC colon. We found this macrophage phenotype is induced upon co-stimulation by IFN-γ and TNF-α collaborating with the Donlin lab.
A proof of concept: This reference can be used to query/investigate cells from other inflammatory diseases and their transcriptomic similarities with our reference which incorporates 5 inflammatory diseased tissues and COVID-19 BALF. We provide a Notebook of example code to show how to map Sepsis PBMCs (Reyes, et al, Nature Medicine, 2020) to our cross-diseased tissue single-cell reference and identify shared inflammatory structures.
Citation: Zhang, Fan, et al. IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Medicine, 2021.