Single-cell transcriptomics in cancer: computational challenges and opportunities. Nature Experimental & Molecular Medicine, 2020
Abstract
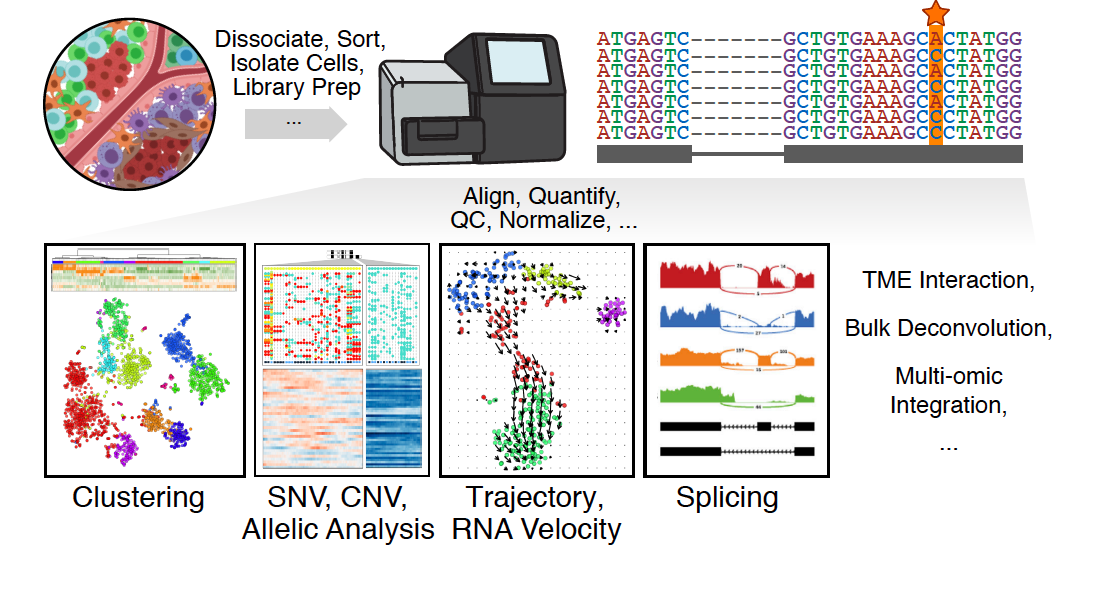
In this review, we highlight emerging themes in computational analysis of single-cell transcriptomics data and their applications to cancer research. We focus on downstream analytical challenges relevant to cancer research, including how to computationally perform unified analysis across many patients and disease states, distinguish neoplastic from non-neoplastic cells, infer communication with the tumor microenvironment, and delineate tumoral and microenvironmental evolution with trajectory and RNA velocity analysis. We include discussions on challenges and opportunities for future computational methodological advancements needed to realize the translational potential of single-cell transcriptomic profiling in cancer.
Type
Publication
Nature Experimental & Molecular Medicine